Registration
Attendance to tutorials is subject to the limitations listed below.
Registration for one or both tutorials can be done through the workshop registration form.
Venue
Tutorials will be held in the Fisciano campus of the University of Salerno.
Both tutorials will be held in room F2, building F2, of the Campus.

Map of the Campus (click to enlarge/download)
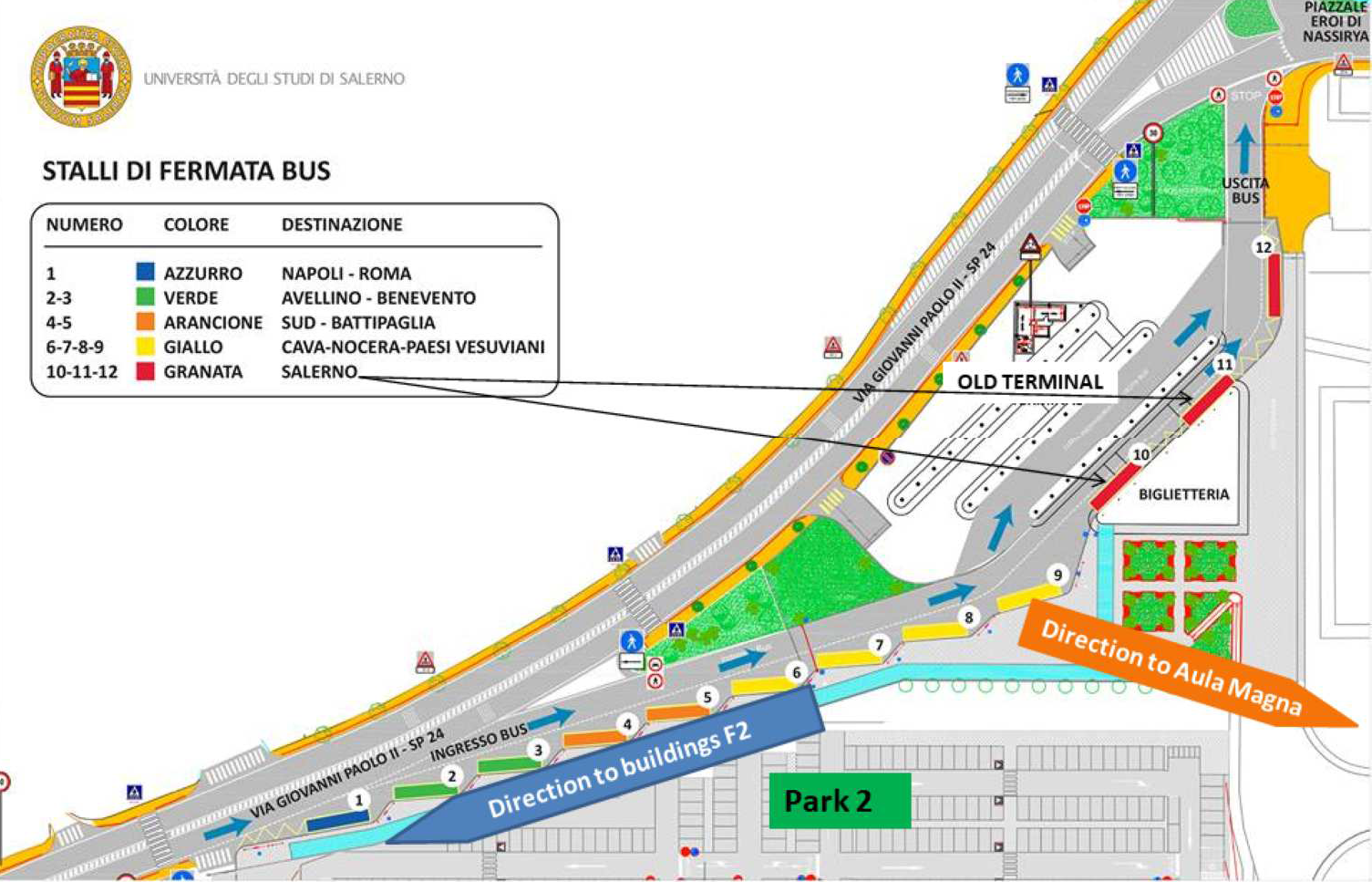
Map of the entrance with directions to venues (click to enlarge/download)
See the document with venue maps and directions for detailed information on how to reach the location of tutorials.
List of tutorials
Tutorial 1, Thursday November 14, 09:00 – 16:00
Analysis of scRNA-seq using R
Annamaria Carissimo, Monika Krzak, Dario Righelli and Claudia Angelini,
Institute for Applied Mathematics “Mauro Picone” (IAC), National Research Council, Naples, Italy.
Registration fee: 10.00 €
Tutorial 2, Monday November 11, 09:30 – 17:30
Label-free quantification with OpenMS
Oliver Kohlbacher, Julianus Pfeuffer and Timo Sachsenberg,
University of Tuebingen, Germany.
Registration fee: 10.00 €

Tutorial supported by the German Network for Bioinformatics Infrastructure.
Tutorial 1
Thursday November 14, 09:00 – 16:00
Analysis of scRNA-seq using R
Provisional Programme
| 09:00-10:00 | Introduction and brief overview on scRNAseq data analysis |
| 10:00-11:20 | Some R packages for creating scRNAseq data object, quality filtering, normalisation, clustering and visualisation: a guided step-by-step example |
| 11:20-11:40 | Coffee break |
| 11:40-13:20 | Group analysis: Participants will be divided in 4 groups. Each group has to to perform a specific data analysis on a given dataset |
| 13:20-14:00 | Lunch break |
| 14:00-15.00 | Group analysis continuation and Group presentation: each group has to make a short 5 minutes presentation of the obtained results |
| 15:00-16:00 | Discussion and an overview of some srRNAseq advanced topics |
Course material will be freely availble as GitHub page
Requirements: Participants need to bring their laptop with R installed (version 3.5 or above). The following R packages need to be installed:
library(scater)
library(M3Drop)
library(monocle)
library(Seurat)
library(mclust)
library(scran)
library(SC3)
For further information, please refer to Annamaria Carissimo
(a.carissimo@na.iac.cnr.it).
Tutorial speakers
 |
 |
 |
 |
| Annamaria Carissimo, IAC/CNR, IT |
Monika Krzak, IAC/CNR, IT |
Dario Righelli, IAC/CNR, IT |
Claudia Angelini, IAC/CNR, IT |
Tutorial 2
Monday November 11, 09:30 – 17:30
Label-free quantification with OpenMS

Tutorial supported by the German Network for Bioinformatics Infrastructure.
Computational mass spectrometry provides important tools and bioinformatic solutions for the analysis of proteomics data. Different methods for label-free quantification have been developed in recent years and were successfully applied in a wide range of studies. Non-targeted methods have shown great potential in unbiased discovery studies.
This de.NBI training event introduces key concepts of label-free analysis using workflow-based processing of real-life data sets. We will introduce several open-source software tools for proteomics, primarily focusing on OpenMS (http://www.OpenMS.org/).
In a hands-on session, we will demonstrate how to combine these tools into complex data analysis workflows including visualization of the results. Participants will have the opportunity to bring their own data and design custom analysis workflows together with instructors.
For participants interested in developing their own algorithms and methods within the OpenMS framework, we provide a brief introduction to pyOpenMS – the python interface to the OpenMS development library.
Training material and handouts will be prepared for both users that want to design proteomic workflows, as well as training material for algorithm and tool developers.
The participants should bring their own laptop computers. Installer versions of required software will be made available. Bringing own data is possible, but not necessary.
Tutorial speakers
 |
 |
 |
|
| Oliver Kohlbacher, University of Tuebingen, DE |
Julianus Pfeuffer, University of Tuebingen, DE |
Timo Sachsenberg, University of Tuebingen, DE |
Julianus Pfeuffer is a Ph.D. student in the field of computational mass spectrometry (bottom-up proteomics). He is one of the core developers of OpenMS.
Timo Sachsenberg is a postdoctoral researcher in the field of computational mass spectrometry. He is one of the core developers of OpenMS.
Oliver Kohlbacher is a professor for applied and translation bioinformatics at University of Tübingen and the University Hospital Tübingen as well as a fellow at the Max Planck Institute for Developmental Biology. His groups are currently focusing on research in computational mass spectrometry, structural bioinformatics, translational bioinformatics, and medical informatics.













